Orchestrate dbt Core projects with Airflow and Cosmos
 dbt Core is an open-source library for analytics engineering that helps users build interdependent SQL models for in-warehouse data transformation, using ephemeral compute of data warehouses. Cosmos is an open-source package developed by Astronomer to run dbt models that are part of a dbt Core project within Airflow.
dbt Core is an open-source library for analytics engineering that helps users build interdependent SQL models for in-warehouse data transformation, using ephemeral compute of data warehouses. Cosmos is an open-source package developed by Astronomer to run dbt models that are part of a dbt Core project within Airflow.
dbt on Airflow with Cosmos and the Astro CLI
The open-source provider package Cosmos allows you to integrate dbt jobs into Airflow by automatically creating Airflow tasks from dbt models. You can turn your dbt Core projects into an Airflow Dag or task group with just a few lines of code.
Why use Airflow with dbt Core?
dbt Core offers the possibility to build modular, reusable SQL components with built-in dependency management and incremental builds.
With Cosmos, you can integrate dbt jobs into your open-source Airflow orchestration environment as standalone Dags or as task groups within Dags.
The benefits of using Airflow with dbt Core include:
- Use Airflow’s data-aware scheduling and Airflow sensors to run models depending on other events in your data ecosystem.
- Turn each dbt model into a task, complete with Airflow features like retries and error notifications, as well as full observability into past runs directly in the Airflow UI.
- Run
dbt teston tables created by individual models immediately after a model has completed. Catch issues before moving downstream and integrate additional data quality checks with your preferred tool to run alongside dbt tests. - Run dbt projects using Airflow connections instead of dbt profiles. You can store all your connections in one place, directly within Airflow or by using a secrets backend.
- Leverage native support for installing and running dbt in a virtual environment to avoid dependency conflicts with Airflow.
- Generate and host dbt docs with Airflow.
With Astro, you get all the above benefits and you can deploy your dbt project to your Astro Deployment independently of your Airflow project using the Astro CLI. For more information, see Deploy dbt projects to Astro.
Time to complete
This tutorial takes approximately 30 minutes to complete.
Assumed knowledge
To get the most out of this tutorial, make sure you have an understanding of:
- The basics of dbt Core. See What is dbt?.
- Airflow fundamentals, such as writing Dags and defining tasks. See Get started with Apache Airflow.
- How Airflow and dbt concepts relate to each other. See Similar dbt & Airflow concepts.
- Airflow operators. See Operators 101.
- Airflow task groups. See Airflow task groups.
- Airflow connections. See Manage connections in Apache Airflow.
Prerequisites
- The Astro CLI.
- Access to a data warehouse supported by dbt Core. See dbt documentation for all supported warehouses. This tutorial uses a Postgres database.
You do not need to have dbt Core installed locally in order to complete this tutorial.
Step 1: Configure your Astro project
To use dbt Core with Airflow install dbt Core in a virtual environment and Cosmos in a new Astro project.
-
Create a new Astro project:
-
Add Cosmos, the Airflow Postgres provider and the dbt Postgres adapter to your Astro project
requirements.txtfile. If you are using a different data warehouse, replaceapache-airflow-providers-postgresanddbt-postgreswith the provider package for your data warehouse. You can find information on all provider packages on the Astronomer registry. -
(Alternative) If you cannot install your dbt adapter in the same environment as Airflow due to package conflicts you can create a dbt executable in a virtual environment. In your
Dockerfileadd the following lines to the end of the file:This code runs a bash command when the Docker image is built that creates a virtual environment called
dbt_venvinside of the Astro CLI scheduler container. Thedbt-postgrespackage, which also containsdbt-core, is installed in the virtual environment. If you are using a different data warehouse, replacedbt-postgreswith the adapter package for your data warehouse.
There are other options to run Cosmos even if you cannot install your dbt adapter in the requirements.txt file or create a virtual environment in your Docker image. See the Cosmos documentation on execution modes for more information.
Step 2: Prepare your dbt project
To integrate your dbt project with Airflow, you need to add the project folder to your Airflow environment. For this step you can either add your own project or follow the steps below to create a simple project using two models.
-
Create a folder called
dbtin yourincludefolder. -
In the
dbtfolder, create a folder calledmy_simple_dbt_project. -
In the
my_simple_dbt_projectfolder add yourdbt_project.yml. This configuration file needs to contain at least the name of the project. This tutorial additionally shows how to inject a variable calledmy_namefrom Airflow into your dbt project. -
Add your dbt models in a subfolder called
modelsin themy_simple_dbt_projectfolder. You can add as many models as you want to run. This tutorial uses the following two models:model1.sql:model2.sql:model1.sqlselects the variablemy_name.model2.sqldepends onmodel1.sqland selects everything from the upstream model.
You should now have the following structure within your Airflow environment:
If storing your dbt project alongside your Airflow project is not feasible, there are other ways to use Cosmos, even if the dbt project is hosted in a different location, for example by using a manifest file to parse the project and a containerized execution mode. See the Cosmos documentation for more information.
Step 3: Create an Airflow connection to your data warehouse
Cosmos allows you to apply Airflow connections to your dbt project.
-
Start Airflow by running
astro dev start. -
In the Airflow UI, go to Admin -> Connections and click +.
-
Create a new connection named
db_conn. Select the connection type and supplied parameters based on the data warehouse you are using. For a Postgres connection, enter the following information:- Connection ID:
db_conn. - Connection Type:
Postgres. - Host: Your Postgres host address.
- Schema: Your Postgres database.
- Login: Your Postgres login username.
- Password: Your Postgres password.
- Port: Your Postgres port.
- Connection ID:
If a connection type for your database isn’t available, you might need to make it available by adding the relevant provider package to requirements.txt and running astro dev restart.
Step 4: Write your Airflow Dag
The Dag you’ll write uses Cosmos to create tasks from existing dbt models and the SQLExecuteQueryOperator to query a table that was created. You can add more upstream and downstream tasks to embed the dbt project within other actions in your data ecosystem.
-
In your
dagsfolder, create a file calledmy_simple_dbt_dag.py. -
Copy and paste the following Dag code into the file:
This Dag uses the
DbtTaskGroupclass from the Cosmos package to create a task group from the models in your dbt project. Dependencies between your dbt models are automatically turned into dependencies between Airflow tasks. Make sure to add your own values forYOUR_NAME,YOUR_DB_NAME, andYOUR_SCHEMA_NAME.Using the
varskeyword in the dictionary provided to theoperator_argsparameter, you can inject variables into the dbt project. This DAG injectsYOUR_NAMEfor themy_namevariable. If your dbt project contains dbt tests, they will be run directly after a model has completed. Note that it is a best practice to setretriesto at least 2 for all tasks that run dbt models.
DagBag import timeout error. This error can be resolved by increasing the value of the Airflow configuration core.dagbag_import_timeout.-
Run the Dag manually by clicking the play button and view the Dag in the graph view. Expand the task groups to see all tasks.
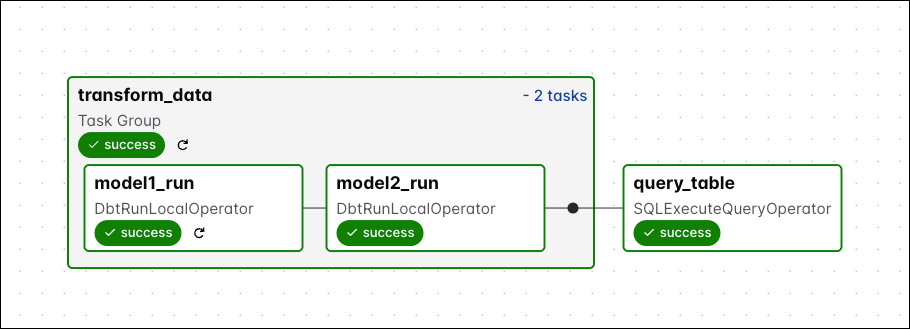
-
Check the XCom returned by the
query_tabletask to see your name in themodel2table.
DbtDag class, as shown in the Cosmos documentation.Congratulations! You’ve run a Dag using Cosmos to automatically create tasks from dbt models. You can learn more about how to configure Cosmos in the Cosmos documentation.
dbt build with the dbt CLI. See the Cosmos documentation for more information.Alternative ways to run dbt Core with Airflow
While using Cosmos is recommended, there are several other ways to run dbt Core with Airflow.
Using the BashOperator
You can use the BashOperator to execute specific dbt commands. It’s recommended to run dbt-core and the dbt adapter for your database in a virtual environment because there often are dependency conflicts between dbt and other packages.
The Dag below uses the BashOperator to activate the virtual environment and execute dbt_run for a dbt project.
Using the BashOperator to run dbt run and other dbt commands can be useful during development. However, running dbt at the project level has a couple of issues:
- There is low observability into what execution state the project is in.
- Failures are absolute and require all models in a project to be run again, which can be costly.
Using a manifest file
Using a dbt-generated manifest.json file gives you more visibility into the steps dbt is running in each task. This file is generated in the target directory of your dbt project and contains its full representation. For more information on this file, see the dbt documentation.
Cosmos can parse manifest files, see the Cosmos documentation for more information.