Orchestrate Cohere LLMs with Apache Airflow
Info
This page has not yet been updated for Airflow 3. The concepts shown are relevant, but some code may need to be updated. If you run any examples, take care to update import statements and watch for any other breaking changes.
Cohere is a natural language processing (NLP) platform that provides an API to access cutting-edge large language models (LLMs). The Cohere Airflow provider offers modules to easily integrate Cohere with Airflow.
In this tutorial, you use Airflow and the Cohere Airflow provider to generate recipe suggestions based on a list of ingredients and countries of recipe origin. Additionally, you create embeddings of the recipes and perform dimensionality reduction using principal component analysis (PCA) to plot recipe similarity in two dimensions.
Why use Airflow with Cohere?
Cohere provides highly specialized out-of-the box and custom LLMs. Countless applications use these models for both user-facing needs, such as to moderate user-generated content, and internal purposes, like providing insight into customer support tickets.
Integrating Cohere with Airflow into one end-to-end machine learning pipeline allows you to:
- Use Airflow’s data-driven scheduling to run operations with Cohere LLM endpoints based on upstream events in your data ecosystem, like when new user input is ingested or a new dataset is available.
- Send several requests to a model endpoint in parallel based on upstream events in your data ecosystem or user input with Airflow params.
- Add Airflow features like retries and alerts to your Cohere operations. This is critical for day 2 MLOps operations, for example, to handle model service outages.
- Use Airflow to orchestrate the creation of vector embeddings with Cohere models, which is especially useful for very large datasets that cannot be processed automatically by vector databases.
Time to complete
This tutorial takes approximately 15 minutes to complete (cooking your recommended recipe not included).
Assumed knowledge
To get the most out of this tutorial, make sure you have an understanding of:
- The basics of the Cohere API. See Cohere Documentation.
- The basics of vector embeddings. See the Cohere Embeddings guide.
- Airflow fundamentals, such as writing DAGs and defining tasks. See Get started with Apache Airflow.
- Airflow operators. See Operators 101.
- Airflow hooks. See Hooks 101.
Prerequisites
- The Astro CLI.
- A Cohere API key. You can generate an API key in the Cohere dashboard, accessible with a Cohere account. A free tier API key is sufficient for this tutorial.
Step 1: Configure your Astro project
-
Create a new Astro project:
-
Add the following lines to your
requirements.txtfile to install the Cohere Airflow provider and other supporting packages: -
To create an Airflow connection to Cohere, add the following environment variables to your
.envfile. Make sure to provide<your-cohere-api-key>.
Step 2: Create your DAG
-
In your
dagsfolder, create a file calledrecipe_suggestions.py. -
Copy the following code into the file.
This DAG consists of five tasks to make a simple MLOps pipeline.
- The
get_ingredientstask fetches the list of ingredients that the user found in their pantry and wants to use in their recipe. The inputpantry_ingredientsparam is provided by Airflow params when you run the DAG. - The
get_countriestask uses Airflow params to retrieve the list of user-provided countries to get recipes from. - The
get_a_recipetask uses the CohereHook to connect to the Cohere API and use the/generateendpoint to get a tasty recipe suggestion based on the user’s pantry ingredients and one of the countries they provided. This task is dynamically mapped over the list of countries to generate one task instance per country. The recipes are saved as.txtfiles in theincludefolder. - The
get_embeddingstask is defined using the CohereEmbeddingOperator to generate vector embeddings of the recipes generated by the upstreamget_a_recipetask. This task is dynamically mapped over the list of recipes to retrieve one set of embeddings per recipe. This pattern allows for efficient parallelization of the vector embedding generation. - The
plot_embeddingstask takes the embeddings created by the upstream task and performs dimensionality reduction using PCA to plot the embeddings in two dimensions.
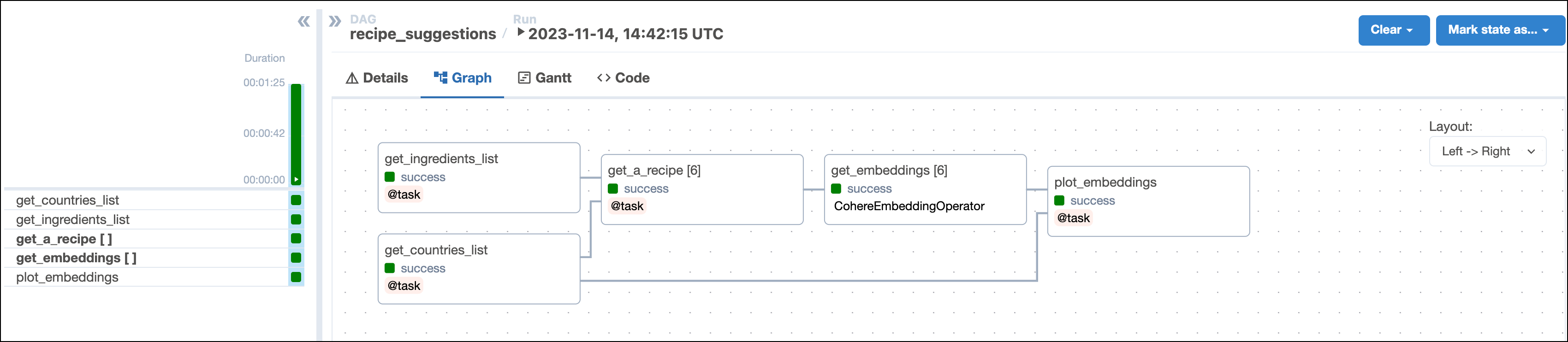
- The
Step 3: Run your DAG
-
Run
astro dev startin your Astro project to start Airflow and open the Airflow UI atlocalhost:8080. -
In the Airflow UI, run the
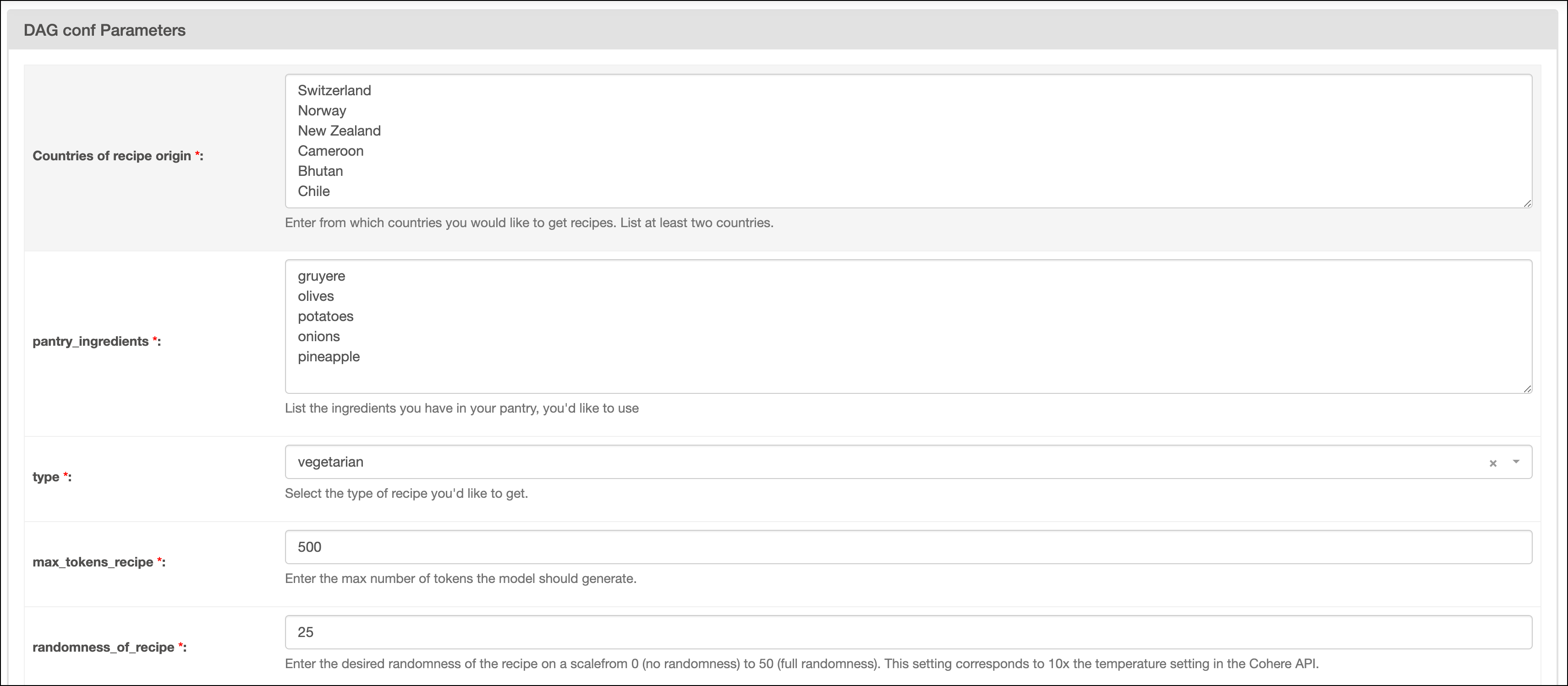
recipe_suggestionsDAG by clicking the play button. Then, provide Airflow params for:Countries of recipe origin: A list of the countries you want to get recipe suggestions from. Make sure to create one line per country and to provide at least two countries.pantry_ingredients: A list of the ingredients you have in your pantry and want to use in the recipe. Make sure to create one line per ingredient.type: Select your preferred recipe type.max_tokens_recipe: The maximum number of tokens available for the recipe.randomness_of_recipe: The randomness of the recipe. The value provided is divided by 10 and given to thetemperatureparameter of the of the Cohere API. The scale for the param ranges from 0 to 50, with 0 being the most deterministic and 50 being the most random.
-
Go to the
includefolder to view the image file created by theplot_embeddingstask. The image should look similar to the one below.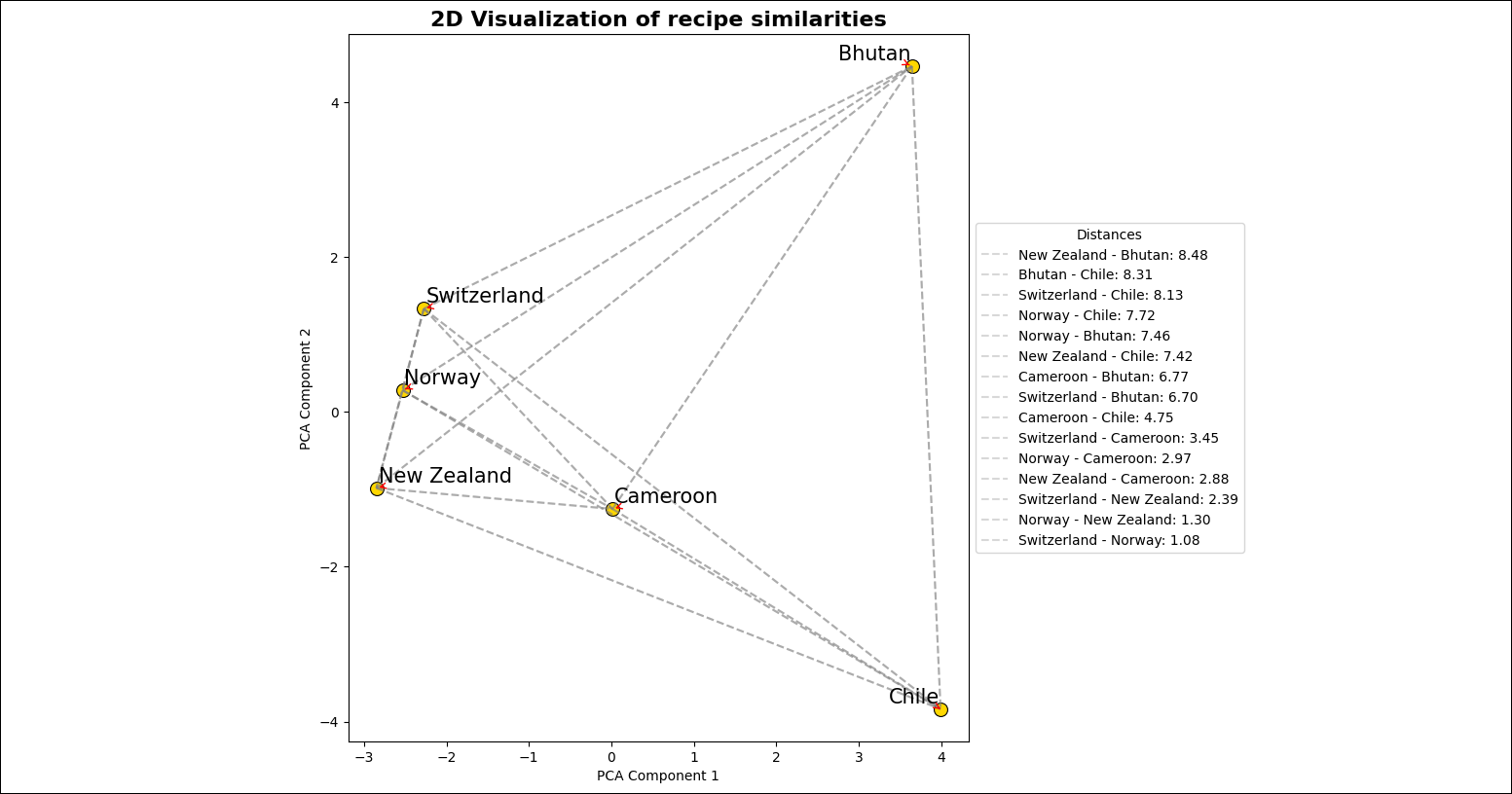
Step 4: (Optional) Cook your recipe
- Choose one of the recipes in the
includefolder. - Navigate to your kitchen and cook the recipe you generated using Cohere with Airflow.
- Enjoy!
Conclusion
Congratulations! You used Airflow and Cohere to get recipe suggestions based on your pantry items. You can now use Airflow to orchestrate Cohere operations in your own machine learning pipelines.