Create dynamic Airflow tasks
With dynamic task mapping, you can write DAGs that dynamically generate parallel tasks at runtime. This feature is a paradigm shift for DAG design in Airflow, since it allows you to create tasks based on the current runtime environment without having to change your DAG code.
In this guide, you'll learn about dynamic task mapping and complete an example implementation for a common use case.
Assumed knowledge
To get the most out of this guide, you should have an understanding of:
- Airflow Operators. See Operators 101.
- How to use Airflow decorators to define tasks. See Introduction to Airflow Decorators.
- XComs in Airflow. See Passing Data Between Airflow Tasks.
Dynamic task concepts
The Airflow dynamic task mapping feature is based on the MapReduce programming model. Dynamic task mapping creates a single task for each input. The reduce procedure, which is optional, allows a task to operate on the collected output of a mapped task. In practice, this means that your DAG can create an arbitrary number of parallel tasks at runtime based on some input parameter (the map), and then if needed, have a single task downstream of your parallel mapped tasks that depends on their output (the reduce).
Airflow tasks have two functions available to implement the map portion of dynamic task mapping. For the task you want to map, you must pass all operator parameters through one of the following functions.
expand(): This function passes the parameters that you want to map. A separate parallel task is created for each input. For some instances of mapping over multiple parameters,.expand_kwargs()is used instead.partial(): This function passes any parameters that remain constant across all mapped tasks which are generated byexpand().
In the following example, the task uses both, .partial() and .expand(), to dynamically generate three task runs.
- TaskFlow API
- Traditional syntax
from airflow.sdk import task
@task
def add(x: int, y: int):
return x + y
added_values = add.partial(y=10).expand(x=[1, 2, 3])
from airflow.providers.standard.operators.python import PythonOperator
def add_function(x: int, y: int):
return x + y
added_values = PythonOperator.partial(
task_id="add",
python_callable=add_function,
op_kwargs={"y": 10},
).expand(op_args=[[1], [2], [3]])

This expand function creates three mapped add tasks, one for each entry in the x input list. The partial function specifies a value for y that remains constant in each task.
When you work with mapped tasks, keep the following in mind:
- You can use the results of an upstream task as the input to a mapped task. The upstream task must return a value in a
dictorlistform. If you're using traditional operators and not decorated tasks, the mapping values must be stored in XComs. - You can map over multiple parameters.
- You can use the results of a mapped task as input to a downstream mapped task.
- You can have a mapped task that results in no task instances. For example, when your upstream task that generates the mapping values returns an empty list. In this case, the mapped task is marked skipped, and downstream tasks are run according to the trigger rules you set. By default, downstream tasks are also skipped.
- Some parameters can't be mapped. For example,
task_id,pool, and manyBaseOperatorarguments. expand()only accepts keyword arguments.- The maximum amount of mapped task instances is determined by the
max_map_lengthparameter in the Airflow configuration. By default it is set to 1024. - You can limit the number of mapped task instances for a particular task that run in parallel by setting the following parameters in your dynamically mapped task:
- Set a limit across all DAG runs with the
max_active_tis_per_dagparameter. - Set a limit for parallel runs within a single DAG with the
max_active_tis_per_dagrunparameter.
- Set a limit across all DAG runs with the
- XComs created by mapped task instances are stored in a list and can be accessed by using the map index of a specific mapped task instance. For example, to access the XComs created by the third mapped task instance (map index of 2) of
my_mapped_task, useti.xcom_pull(task_ids=['my_mapped_task'])[2]. Themap_indexesparameter in the.xcom_pull()method allows you to specify a list of map indexes of interest (ti.xcom_pull(task_ids=['my_mapped_task'], map_indexes=[2])).
For additional examples of how to apply dynamic task mapping functions, see Dynamic Task Mapping in the official Airflow documentation.
The Airflow UI provides observability for mapped tasks in the Grid View.
Mapped tasks are identified with a set of brackets [ ] followed by the task ID. All mapped task instances are combined into one row on the grid.
The number in the brackets show in the DAG run graph, is updated for each DAG run to reflect how many mapped instances were created. The following screenshot shows a DAG run graph with two tasks, the latter having 49 dynamically mapped task instances.
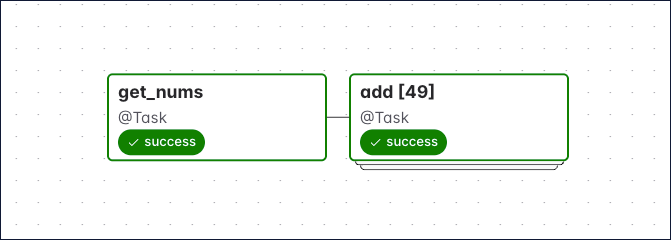
To see the logs for and XCom pushed by each dynamically mapped task instance, click on on the grid square of a dynamically mapped task and then on an individual task instance.
Mapping over the result of another operator
You can use the output of an upstream operator as the input data for a dynamically mapped downstream task.
In this section you'll learn how to pass mapping information to a downstream task for each of the following scenarios:
- TaskFlow over TaskFlow: Both tasks are defined using the TaskFlow API.
- TaskFlow over traditional operator: The upstream task is defined using a traditional operator and the downstream task is defined using the TaskFlow API.
- Traditional operator over TaskFlow: The upstream task is defined using the TaskFlow API and the downstream task is defined using a traditional operator.
- Traditional operator over traditional operator: Both tasks are defined using traditional operators.
- TaskFlow over TaskFlow
- TaskFlow over Traditional operator
- Traditional operator over TaskFlow
- Traditional operator over Traditional operator
If both tasks are defined using the TaskFlow API, you can provide a function call to the upstream task as the argument for the expand() function.
from airflow.sdk import task
@task
def one_two_three_TF():
return [1, 2, 3]
@task
def plus_10_TF(x):
return x + 10
plus_10_TF.partial().expand(x=one_two_three_TF())
If you are mapping over the results of a traditional operator, you need to provide the argument for expand() using the .output attribute of the task object.
from airflow.sdk import task
from airflow.providers.standard.operators.python import PythonOperator
def one_two_three_traditional():
return [1, 2, 3]
@task
def plus_10_TF(x):
return x + 10
one_two_three_task = PythonOperator(
task_id="one_two_three_task", python_callable=one_two_three_traditional
)
plus_10_TF.partial().expand(x=one_two_three_task.output)
When mapping a traditional PythonOperator over results from an upstream TaskFlow task you need to modify the format of the output to be accepted by the op_args argument of the traditional PythonOperator.
from airflow.sdk import task
from airflow.providers.standard.operators.python import PythonOperator
@task
def one_two_three_TF():
# this adjustment is due to op_args expecting each argument as a list
return [[1], [2], [3]]
def plus_10_traditional(x):
return x + 10
plus_10_task = PythonOperator.partial(
task_id="plus_10_task", python_callable=plus_10_traditional
).expand(op_args=one_two_three_TF())
When mapping a traditional PythonOperator over the result of another PythonOperator use the .output attribute on the task object and make sure the format returned by the upstream task matches the format expected by the op_args parameter.
from airflow.providers.standard.operators.python import PythonOperator
def one_two_three_traditional():
# this adjustment is due to op_args expecting each argument as a list
return [[1], [2], [3]]
def plus_10_traditional(x):
return x + 10
one_two_three_task = PythonOperator(
task_id="one_two_three_task", python_callable=one_two_three_traditional
)
plus_10_task = PythonOperator.partial(
task_id="plus_10_task", python_callable=plus_10_traditional
).expand(op_args=one_two_three_task.output)
# when only using traditional operators, define dependencies explicitly
one_two_three_task >> plus_10_task
Map over combined results of upstream tasks
You can combine the output lists of upstream tasks using the .concat() method. In previous Airflow versions, you needed an intermediate task combining the lists to achieve this.
The code snippet shown below creates 7 dynamically mapped task instances for the map_me task.
- .concat() with TaskFlow
- Traditional operator over Traditional operator
from airflow.sdk import task
import time
@task
def t1():
return [1, 2, 3]
t1_obj = t1()
@task
def t2():
return [4, 5, 6, 7]
t2_obj = t2()
@task
def map_me(input):
print(f"Sleeping for {input} seconds!")
time.sleep(input)
print("Waking up!")
map_me.expand(input=t1_obj.concat(t2_obj))
from airflow.providers.standard.operators.python import PythonOperator
import time
def t1_func():
return [[1], [2], [3]]
def t2_func():
return [[4], [5], [6], [7]]
def map_me_func(input):
print(f"Sleeping for {input} seconds!")
time.sleep(input)
print("Waking up!")
t1 = PythonOperator(task_id="t1", python_callable=t1_func)
t2 = PythonOperator(task_id="t2", python_callable=t2_func)
map_me = PythonOperator.partial(
task_id="map_me", python_callable=map_me_func
).expand(op_args=t1.output.concat(t2.output))
Mapping over multiple parameters
You can use one of the following methods to map over multiple parameters:
- Cross-product: Mapping over two or more keyword arguments results in a mapped task instance for each possible combination of inputs. This type of mapping uses the
expand()function. - Sets of keyword arguments: Mapping over two or more sets of one or more keyword arguments results in a mapped task instance for every defined set, rather than every combination of individual inputs. This type of mapping uses the
expand_kwargs()function. - Zip: Mapping over a set of positional arguments created with Python's built-in
zip()function or with the.zip()method of an XComArg results in one mapped task for every set of positional arguments. Each set of positional arguments is passed to the same keyword argument of the operator. This type of mapping uses theexpand()function.
Cross-product
The default behavior of the expand() function is to create a mapped task instance for every possible combination of all provided inputs. For example, if you map over three keyword arguments and provide two options to the first, four options to the second, and five options to the third, you would create 2x4x5=40 mapped task instances. One common use case for this method is tuning model hyperparameters.
The following task definition maps over three options for the bash_command parameter and three options for the env parameter. This will result in 3x3=9 mapped task instances. Each bash command runs with each definition for the environment variable WORD.
from airflow.providers.standard.operators.bash import BashOperator
cross_product_example = BashOperator.partial(
task_id="cross_product_example"
).expand(
bash_command=[
"echo $WORD", # prints the env variable WORD
"echo `expr length $WORD`", # prints the number of letters in WORD
"echo ${WORD//e/X}" # replaces each "e" in WORD with "X"
],
env=[
{"WORD": "hello"},
{"WORD": "tea"},
{"WORD": "goodbye"}
]
)
The nine mapped task instances of the task cross_product_example run all possible combinations of the bash command with the env variable:
- Map index 0:
hello - Map index 1:
tea - Map index 2:
goodbye - Map index 3:
5 - Map index 4:
3 - Map index 5:
7 - Map index 6:
hXllo - Map index 7:
tXa - Map index 8:
goodbyX
Sets of keyword arguments
To map over sets of inputs to two or more keyword arguments (kwargs), you can use the expand_kwargs() function. You can provide sets of parameters as a list containing a dictionary or as an XComArg. The operator gets 3 sets of commands, resulting in 3 mapped task instances.
from airflow.providers.standard.operators.bash import BashOperator
# input sets of kwargs provided directly as a list[dict]
t1 = BashOperator.partial(task_id="t1").expand_kwargs(
[
{"bash_command": "echo $WORD", "env" : {"WORD": "hello"}},
{"bash_command": "echo `expr length $WORD`", "env" : {"WORD": "tea"}},
{"bash_command": "echo ${WORD//e/X}", "env" : {"WORD": "goodbye"}}
]
)
The task t1 will have three mapped task instances printing their results into the logs:
- Map index 0:
hello - Map index 1:
3 - Map index 2:
goodbyX
Zip
In dynamic task mapping, you can provide sets of positional arguments to the same keyword argument. For example, the op_args argument of the PythonOperator. You can use the built-in zip() Python function if your inputs are in the form of iterables such as tuples, dictionaries, or lists. If your inputs come from XCom objects, you can use the .zip() method of the XComArg object.
Provide positional arguments with the built-in Python zip()
The zip() function takes in an arbitrary number of iterables and uses their elements to create a zip-object containing tuples. There will be as many tuples as there are elements in the shortest iterable. Each tuple contains one element from every iterable provided. For example:
zip(["a", "b", "c"], [1, 2, 3], ["hi", "bye", "tea"])results in a zip object containing:("a", 1, "hi"), ("b", 2, "bye"), ("c", 3, "tea").zip(["a", "b"], [1], ["hi", "bye"], [19, 23], ["x", "y", "z"])results in a zip object containing only one tuple:("a", 1, "hi", 19, "x"). This is because the shortest list provided only contains one element.- It is also possible to zip together different types of iterables.
zip(["a", "b"], {"hi", "bye"}, (19, 23))results in a zip object containing:('a', 'hi', 19), ('b', 'bye', 23).
The following code snippet shows how a list of zipped arguments can be provided to the expand() function in order to create mapped tasks over sets of positional arguments. In the TaskFlow API version of the DAG, each set of positional arguments is passed to the argument zipped_x_y_z. In the DAG using a traditional PythonOperator each set of positional arguments is unpacked due to op_args expecting an iterable and passed to the arguments x, y and z.
- TaskFlow API
- Traditional syntax
from airflow.sdk import task
# use the zip function to create three-tuples out of three lists
zipped_arguments = list(zip([1, 2, 3], [10, 20, 30], [100, 200, 300]))
# zipped_arguments contains: [(1,10,100), (2,20,200), (3,30,300)]
# creating the mapped task instances using the TaskFlow API
@task
def add_numbers(zipped_x_y_z):
return zipped_x_y_z[0] + zipped_x_y_z[1] + zipped_x_y_z[2]
add_numbers.expand(zipped_x_y_z=zipped_arguments)
from airflow.providers.standard.operators.python import PythonOperator
# use the zip function to create three-tuples out of three lists
zipped_arguments = list(zip([1, 2, 3], [10, 20, 30], [100, 200, 300]))
# zipped_arguments contains: [(1,10,100), (2,20,200), (3,30,300)]
# function for the PythonOperator
def add_numbers_function(x, y, z):
return x + y + z
# dynamically mapped PythonOperator
add_numbers = PythonOperator.partial(
task_id="add_numbers",
python_callable=add_numbers_function,
).expand(op_args=zipped_arguments)
The task add_numbers will have three mapped task instances one for each tuple of positional arguments:
- Map index 0:
111 - Map index 1:
222 - Map index 2:
333
Provide positional arguments with XComArg.zip()
It is also possible to zip XComArg objects. If the upstream task has been defined using the TaskFlow API, provide the function call. If the upstream task uses a traditional operator, provide task_object.output or XcomArg(task_object). In the following example, you can see the results of three tasks being zipped together to form the zipped_arguments ([(1, 10, 100), (2, 1000, 200), (1000, 1000, 300)]).
To mimic the behavior of the zip_longest() function, you can add the optional fillvalue keyword argument to the .zip() method. If you specify a default value with fillvalue, the method produces as many tuples as the longest input has elements and fills in missing elements with the default value. If fillvalue was not specified in the example below, zipped_arguments would only contain one tuple [(1, 10, 100)] since the shortest list provided to the .zip() method is only one element long.
- TaskFlow API
- Traditional syntax
from airflow.sdk import task
@task
def one_two_three():
return [1, 2]
@task
def ten_twenty_thirty():
return [10]
@task
def one_two_three_hundred():
return [100, 200, 300]
zipped_arguments = one_two_three().zip(
ten_twenty_thirty(), one_two_three_hundred(), fillvalue=1000
)
# zipped_arguments contains [(1, 10, 100), (2, 1000, 200), (1000, 1000, 300)]
# creating the mapped task instances using the TaskFlow API
@task
def add_nums(zipped_x_y_z):
return zipped_x_y_z[0] + zipped_x_y_z[1] + zipped_x_y_z[2]
add_nums.expand(zipped_x_y_z=zipped_arguments)
from airflow.providers.standard.operators.python import PythonOperator
def one_two_three_function():
return [1, 2]
def ten_twenty_thirty_function():
return [10]
def one_two_three_hundred_function():
return [100, 200, 300]
one_two_three = PythonOperator(
task_id="one_two_three", python_callable=one_two_three_function
)
ten_twenty_thirty = PythonOperator(
task_id="ten_twenty_thirty", python_callable=ten_twenty_thirty_function
)
one_two_three_hundred = PythonOperator(
task_id="one_two_three_hundred", python_callable=one_two_three_hundred_function
)
zipped_arguments = one_two_three.output.zip(
ten_twenty_thirty.output, one_two_three_hundred.output, fillvalue=1000
)
# zipped_arguments contains [(1, 10, 100), (2, 1000, 200), (1000, 1000, 300)]
# function that will be used in the dynamically mapped PythonOperator
def add_nums_function(x, y, z):
return x + y + z
add_nums = PythonOperator.partial(
task_id="add_nums", python_callable=add_nums_function
).expand(op_args=zipped_arguments)
The add_nums task will have three mapped instances with the following results:
- Map index 0:
111(1+10+100) - Map index 1:
1202(2+1000+200) - Map index 2:
2300(1000+1000+300)
Repeated mapping
You can dynamically map an Airflow task over the output of another dynamically mapped task. This results in one mapped task instance for every mapped task instance of the upstream task.
The following example shows three dynamically mapped tasks.
- TaskFlow API
- Traditional syntax
from airflow.sdk import task
@task
def multiply_by_2(num):
return num * 2
@task
def add_10(num):
return num + 10
@task
def multiply_by_100(num):
return num * 100
multiplied_value_1 = multiply_by_2.expand(num=[1, 2, 3])
summed_value = add_10.expand(num=multiplied_value_1)
multiply_by_100.expand(num=summed_value)
from airflow.providers.standard.operators.python import PythonOperator
def multiply_by_2_func(num):
return [num * 2]
def add_10_func(num):
return [num + 10]
def multiply_by_100_func(num):
return num * 100
multiply_by_2 = PythonOperator.partial(
task_id="multiply_by_2",
python_callable=multiply_by_2_func
).expand(op_args=[[1], [2], [3]])
add_10 = PythonOperator.partial(
task_id="add_10",
python_callable=add_10_func
).expand(op_args=multiply_by_2.output)
multiply_by_100 = PythonOperator.partial(
task_id="multiply_by_100",
python_callable=multiply_by_100_func
).expand(op_args=add_10.output)
multiply_by_2 >> add_10 >> multiply_by_100
In the example above, the multiply_by_2 task is dynamically mapped over a list of three elements ([1, 2, 3]). The task has three mapped task instances containing the following values:
- Map index 0:
2(1*2) - Map index 1:
4(2*2) - Map index 2:
6(3*2)
The add_10 task is dynamically mapped over the output of the multiply_by_2 task. It has 3 mapped task instances (one for each mapped instance of the previous task) which contain the following values:
- Map index 0:
12(2+10) - Map index 1:
14(4+10) - Map index 2:
16(6+10)
The multiply_by_100 task is dynamically mapped over the output of the add_10 task, which results in three mapped task instances with the following outputs:
- Map index 0:
1200(12*100) - Map index 1:
1400(14*100) - Map index 2:
1600(16*100)
You can chain an arbitrary number of dynamically mapped tasks in this manner. It is currently not possible to exponentially increase the number of mapped task instances.
Mapping over task groups
Task groups defined with the @task_group decorator can be dynamically mapped as well. The syntax for dynamically mapping over a task group is the same as dynamically mapping over a single task.
from airflow.sdk import task, task_group
# creating a task group using the decorator with the dynamic input my_num
@task_group(group_id="group1")
def tg1(my_num):
@task
def print_num(num):
return num
@task
def add_42(num):
return num + 42
print_num(my_num) >> add_42(my_num)
# creating 6 mapped task group instances of the task group group1
tg1_object = tg1.expand(my_num=[19, 23, 42, 8, 7, 108])
You can also dynamically map over multiple task group input parameters as you would for regular tasks using a cross-product, zip function, or sets of keyword arguments. For more on this, see Mapping over multiple parameters.
Transform outputs with .map
There are use cases where you want to transform the output of an upstream task before another task dynamically maps over it. For example, if the upstream traditional operator returns its output in a fixed format or if you want to skip certain mapped task instances based on a logical condition.
The .map() method accepts a Python function and uses it to transform an iterable input before a task dynamically maps over it.
You can call .map() directly on a task using the TaskFlow API (my_upstream_task_flow_task().map(mapping_function)) or on the output object of a traditional operator (my_upstream_traditional_operator.output.map(mapping_function)).
The downstream task is dynamically mapped over the object created by the .map() method using either .expand() for a single keyword argument or .expand_kwargs() for list of dictionaries containing sets of keyword arguments.
The code snippet below shows how to use .map() to skip specific mapped tasks based on a logical condition.
list_stringsis the upstream task returning a list of strings. -skip_strings_starting_with_skiptransforms a list of strings into a list of modified strings andAirflowSkipExceptions. In this DAG, the function transformslist_stringsinto a new list calledtransformed_list. This function will not appear as an Airflow task.mapped_printing_taskdynamically maps over thetransformed_listobject.
- TaskFlow API
- Traditional syntax
from airflow.sdk import task
from airflow.exceptions import AirflowSkipException
# an upstream task returns a list of outputs in a fixed format
@task
def list_strings():
return ["skip_hello", "hi", "skip_hallo", "hola", "hey"]
# the function used to transform the upstream output before
# a downstream task is dynamically mapped over it
def skip_strings_starting_with_skip(string):
if len(string) < 4:
return string + "!"
elif string[:4] == "skip":
raise AirflowSkipException(f"Skipping {string}; as I was told!")
else:
return string + "!"
# transforming the output of the first task with the map function.
transformed_list = list_strings().map(skip_strings_starting_with_skip)
# the task using dynamic task mapping on the transformed list of strings
@task
def mapped_printing_task(string):
return "Say " + string
mapped_printing_task.partial().expand(string=transformed_list)
from airflow.providers.standard.operators.python import PythonOperator
from airflow.exceptions import AirflowSkipException
# an upstream task returns a list of outputs in a fixed format
def list_strings():
return ["skip_hello", "hi", "skip_hallo", "hola", "hey"]
listed_strings = PythonOperator(
task_id="list_strings",
python_callable=list_strings,
)
# the function used to transform the upstream output before
# a downstream task is dynamically mapped over it
def skip_strings_starting_with_skip(string):
if len(string) < 4:
return [string + "!"]
elif string[:4] == "skip":
raise AirflowSkipException(f"Skipping {string}; as I was told!")
else:
return [string + "!"]
# transforming the output of the first task with the map function.
# since `op_args` expects a list of lists it is important
# each element of the list is wrapped in a list in the map function.
transformed_list = listed_strings.output.map(skip_strings_starting_with_skip)
# function to use in the dynamically mapped PythonOperator
def mapped_printing_function(string):
return "Say " + string
mapped_printing = PythonOperator.partial(
task_id="mapped_printing",
python_callable=mapped_printing_function,
).expand(op_args=transformed_list)
In the [] Mapped Tasks tab, you can see how the mapped task instances 0 and 2 have been skipped.

Example implementation
For this example, you'll implement one of the most common use cases for dynamic tasks: processing files in Amazon S3. In this scenario, you'll use an ELT framework to extract data from files in Amazon S3, load the data into Snowflake, and transform the data using Snowflake's built-in compute. It's assumed that the files will be dropped daily, but it's unknown how many will arrive each day. You'll leverage dynamic task mapping to create a unique task for each file at runtime. This gives you the benefit of atomicity, better observability, and easier recovery from failures.
All code used in this example is located in the dynamic-task-mapping-tutorial repository.
The example DAG completes the following steps:
- Use a decorated Python operator to get the current list of files from Amazon S3. The Amazon S3 prefix passed to this function is parameterized with
ds_nodashso it pulls files only for the execution date of the DAG run. For example, for a DAG run on April 12th 2025, you assume the files landed in a folder named20250412/. - Use the results of the first task, map an
S3ToSnowflakeOperatorfor each file. - Move the daily folder of processed files into a
processed/folder while, - Simultaneously runs a Snowflake query that transforms the data. The query is located in a separate SQL file in our
include/directory. - Deletes the folder of daily files now that it has been moved to
processed/for record keeping.
- TaskFlow API
- Traditional syntax
from airflow.sdk import dag, task
from airflow.providers.snowflake.transfers.copy_into_snowflake import (
CopyFromExternalStageToSnowflakeOperator,
)
from airflow.providers.snowflake.operators.snowflake import SnowflakeOperator
from airflow.providers.amazon.aws.hooks.s3 import S3Hook
from airflow.providers.amazon.aws.operators.s3 import S3CopyObjectOperator
from airflow.providers.amazon.aws.operators.s3 import S3DeleteObjectsOperator
@dag(template_searchpath="/usr/local/airflow/include")
def mapping_elt():
@task
def get_s3_files(current_prefix):
s3_hook = S3Hook(aws_conn_id="s3")
current_files = s3_hook.list_keys(
bucket_name="my-bucket",
prefix=current_prefix + "/",
start_after_key=current_prefix + "/",
)
return [[file] for file in current_files]
copy_to_snowflake = CopyFromExternalStageToSnowflakeOperator.partial(
task_id="load_files_to_snowflake",
stage="MY_STAGE",
table="COMBINED_HOMES",
schema="MYSCHEMA",
file_format="(type = 'CSV',field_delimiter = ',', skip_header=1)",
snowflake_conn_id="snowflake",
).expand(files=get_s3_files(current_prefix="{{ ds_nodash }}"))
move_s3 = S3CopyObjectOperator(
task_id="move_files_to_processed",
aws_conn_id="s3",
source_bucket_name="my-bucket",
source_bucket_key="{{ ds_nodash }}" + "/",
dest_bucket_name="my-bucket",
dest_bucket_key="processed/" + "{{ ds_nodash }}" + "/",
)
delete_landing_files = S3DeleteObjectsOperator(
task_id="delete_landing_files",
aws_conn_id="s3",
bucket="my-bucket",
prefix="{{ ds_nodash }}" + "/",
)
transform_in_snowflake = SnowflakeOperator(
task_id="run_transformation_query",
sql="/transformation_query.sql",
snowflake_conn_id="snowflake",
)
copy_to_snowflake >> [move_s3, transform_in_snowflake]
move_s3 >> delete_landing_files
mapping_elt()
from airflow.sdk import DAG
from airflow.providers.snowflake.transfers.copy_into_snowflake import (
CopyFromExternalStageToSnowflakeOperator,
)
from airflow.providers.standard.operators.python import PythonOperator
from airflow.providers.snowflake.operators.snowflake import SnowflakeOperator
from airflow.providers.amazon.aws.hooks.s3 import S3Hook
from airflow.providers.amazon.aws.operators.s3 import S3CopyObjectOperator
from airflow.providers.amazon.aws.operators.s3 import S3DeleteObjectsOperator
def get_s3_files(current_prefix):
s3_hook = S3Hook(aws_conn_id="s3")
current_files = s3_hook.list_keys(
bucket_name="my-bucket",
prefix=current_prefix + "/",
start_after_key=current_prefix + "/",
)
return [[file] for file in current_files]
with DAG(
"mapping_elt_traditional",
template_searchpath="/usr/local/airflow/include",
):
get_s3_files_task = PythonOperator(
task_id="get_s3_files",
python_callable=get_s3_files,
op_kwargs={"current_prefix": "{{ ds_nodash }}"},
)
copy_to_snowflake = CopyFromExternalStageToSnowflakeOperator.partial(
task_id="load_files_to_snowflake",
stage="MY_STAGE",
table="COMBINED_HOMES",
schema="MYSCHEMA",
file_format="(type = 'CSV',field_delimiter = ',', skip_header=1)",
snowflake_conn_id="snowflake",
).expand(files=get_s3_files_task.output)
move_s3 = S3CopyObjectOperator(
task_id="move_files_to_processed",
aws_conn_id="s3",
source_bucket_name="my-bucket",
source_bucket_key="{{ ds_nodash }}" + "/",
dest_bucket_name="my-bucket",
dest_bucket_key="processed/" + "{{ ds_nodash }}" + "/",
)
delete_landing_files = S3DeleteObjectsOperator(
task_id="delete_landing_files",
aws_conn_id="s3",
bucket="my-bucket",
prefix="{{ ds_nodash }}" + "/",
)
transform_in_snowflake = SnowflakeOperator(
task_id="run_transformation_query",
sql="/transformation_query.sql",
snowflake_conn_id="snowflake",
)
copy_to_snowflake >> [move_s3, transform_in_snowflake]
move_s3 >> delete_landing_files
The graph of this DAG looks similar to this image:

When dynamically mapping tasks, make note of the format needed for the parameter you are mapping. In the previous example, you wrote your own Python function to get the Amazon S3 keys because the S3toSnowflakeOperator requires each s3_key parameter to be in a list format, and the s3_hook.list_keys function returns a single list with all keys. By writing your own simple function, you can turn the hook results into a list of lists that can be used by the downstream operator.